Portfolio
select research & Design projects
Living Artifacts
biotic design
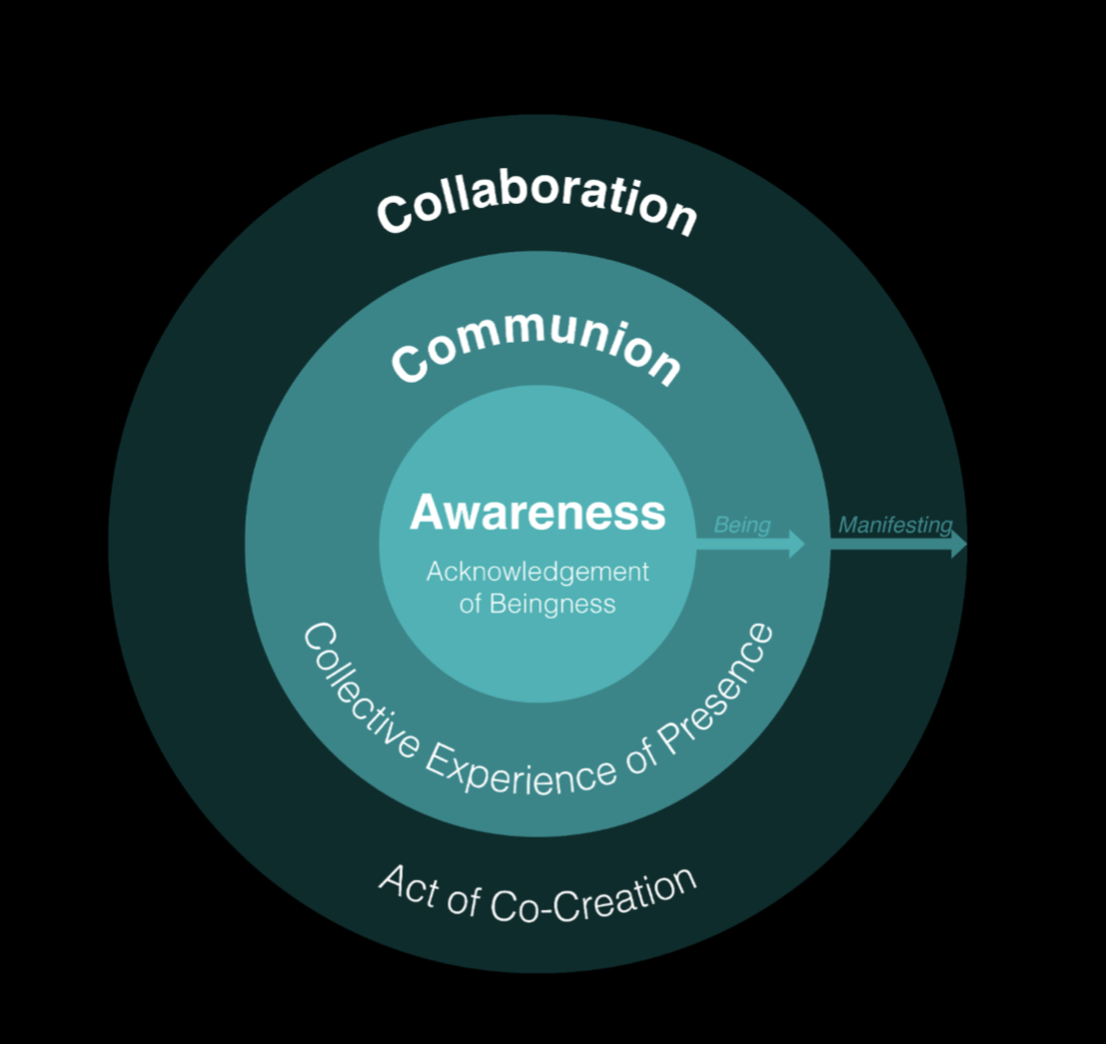
Through Living Artifacts, humanity can explore its relationship with the living world through different levels of conscious engagement: first, through Awareness—an acknowledgement of beingness; then, through Communion, a collective experience of presence; and finally, via Collaboration, an act of co-creation.
Examples of Living Artifacts include the exploration of ‘Living Time’—clocks and other time pieces that reveal time through changes in odor and color, but at the time scales of invisible micro-organisms; ‘Living Cycle’—visualizing the cycles of life and death and renewal mediated by mycelium; and ‘Bacterial biocement,’ a collaboration with microbes to fabricate structures.
Bacterial and Fungal Biocement
engineered living materials
Bacterial biocement is a living material that offers a path to address the repercussions of the built environment while also transforming how we interact with buildings over their lifespan, all made possible through a collaboration with these microscopic living organisms.
Living brick modules were grown with biocementing bacteria. This engineered living material has the potential to mitigate the excessive use of concrete. The bacteria remain alive in the bricks and can communicate with colors in response to environmental factors such as pollutants, helping to map our eco-health. This process of biocementation results in living materials that can address both the repercussions and our perception of the built environment, transforming buildings from static entities situated in a landscape, to living systems integrated within their ecosystems.
In collaboration with Professor Chris Voigt of the Biological Engineering department at MIT, fungal living materials are also being explored.
Field Guide for Growing Community Power
cultivating creativity globally
The world’s coral reefs are dying. Without urgent intervention, our ocean ecosystems are in danger of suffering extraordinary loss of biodversity and other deleterious impacts.
In collaboration with Revive & Restore and Parley for the Ocean, we are developing a “Field Guide for Growing Community Power,” to enable local communities and stakeholders to grow “power-with,” and expand their creative, social, and technical capacities to deploy critical interventions and help enhance the resiliency of corals. From coral translocation to the deployment of coral probiotics, numerous methods are being developed to augment corals in increasingly threatening environmental conditions. Using methods developed in our Field Guide, stakeholders in government, conservation science, and local communities can explore their shared values and develop action plans to deliver urgently needed interventions to help ocean corals overcome the myriad threats to their survival.
Zap-pore
low-cost open-source hardware
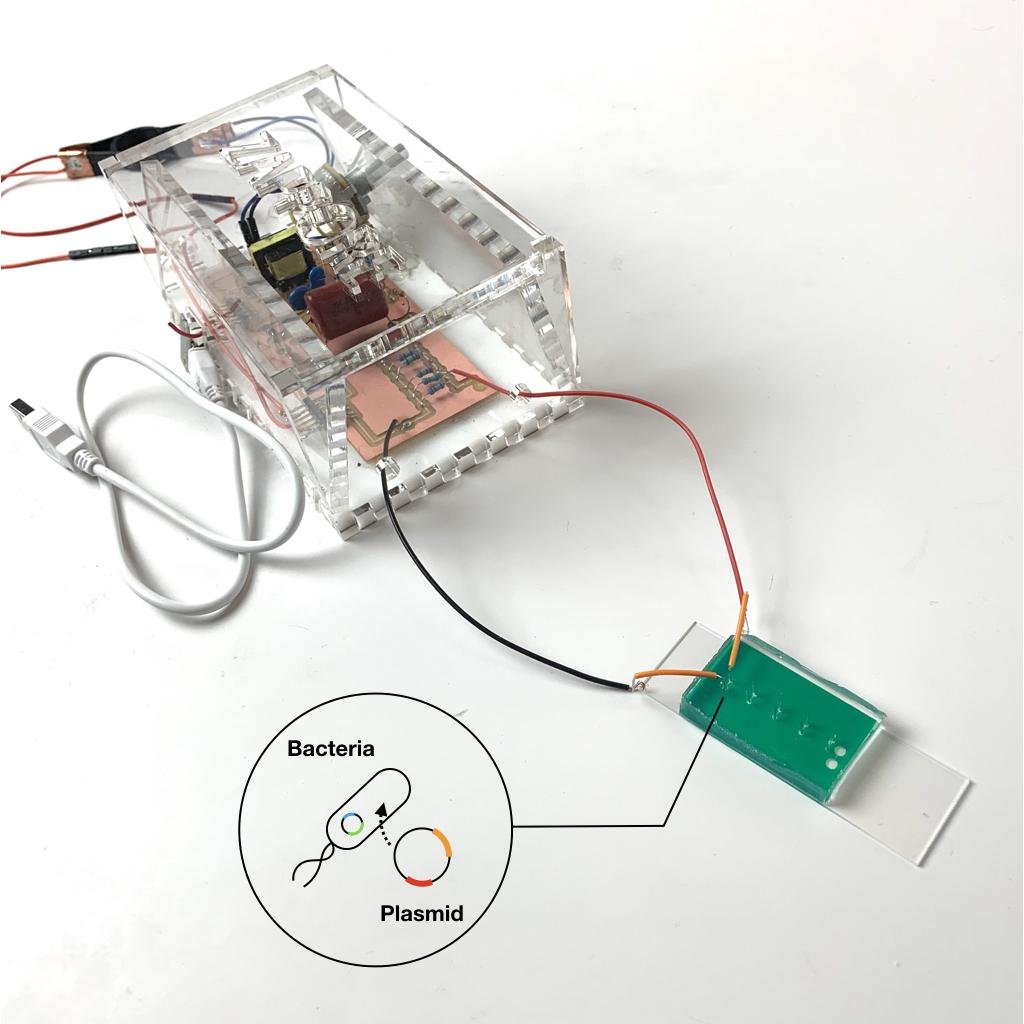
Transformation is fundamental technique in life sciences: the insertion of foreign DNA, in the form of circular DNA strands (known as plasmids), into microbes. Transformation is a critical step in expressing bio molecules of interest in microbes. Transformations are commonly conducted using an electroporator, where electric voltage is applied to generate pores in cell membranes, which enable uptake of DNA into the cell.
Zap-pore is an open-source, low-cost ($10 material bill), DIY tool enabling users to perform transformations using a variety of voltages and pulse times. Zap-pore was designed to be built-in resource-constrained settings. It is a tool designed to explore and experiment the possibility of transformation for known and unknown strains of microbes.
Co-Culture
open-source tool for studying microbiome dynamics
We aim to create an easy-to-use, scalable, widely accessible method to allow for the co-culturing of multiple organisms through the use of water droplets stabilized by surfactants in oil.
This methodology will be used to explore and study the effects of various strains of biota living in the human microbiome upon each other.
Pandemic Response CoLab
collective intelligence
Pandemic Response CoLab is a project of the MIT Center for Collective Intelligence, the MIT Media Lab Community Biotechnology Initiative and Millipore Sigma, building upon a decade of work on MIT Climate CoLab, a platform where more than 120,000 people are working together to develop novel ideas to address climate change.
The Pandemic Response CoLab helped individuals and groups work together to solve practical problems created by the COVID-19 pandemic. By leveraging an open online collaboration platform, we mobilized innovators, communities, businesses, and others to develop actionable solutions to real problems.
Biota Beats
BIOTIC DESIGN
We have built Biota Beats, a microbial record player that translates data about microbes of the body and environment into sound and music. Microbes from the human microbiota, the ecosystem of micro-organisms that inhabit the body, are cultured on custom biota records in our incubator system. Data gathered from the microbes are then sonified to produce beats from the bacteria of the body. Tune into the music of your microbes! Biota Beats has been exhibited at the Stedelijik Museum Breda, the New Jersey Liberty Science Center, and the International Genetically Engineered Machines Competition.
Metafluidics
open repository for bio-hardware
Fluidics are promising foundational tools for synthetic biology. Unfortunately, fluidics are not broadly used, because they are difficult to manufacture and operate, and designs are currently not shared in a systematic fashion. Metafluidics is an open repository of both device and hardware designs to enable communities of users from around the world to share and remix bio-hardware. This repository is the hardware portal for the National Science Foundation supported Living Computing Project. See our Nature Biotechnology paper here. Share your fluidic devices with the global community today!
Artificial Gut
synthetic organs
The human gut microbiota is one of the most densely populated ecosystems of microorganisms on earth. With an estimated 100 trillion micro-organisms, the gut is an extraordinarily complex system of microbe-microbe and microbe-host interactions. A project of the MIT Lincoln Laboratory in collaboration with Professor Eric Alm, co-director of the MIT Microbiome Center, we are building a three-dimensional, artificial gut using the latest digital fabrication technologies for the systematic construction and study of synthetic microbial communities, including exploring the influence of engineered microorganisms on population dynamics.
3D Printed Fluidic Hardware for DNA Assembly
synbio | digital fabrication
The ability to rapidly and cheaply produce hardware for biological engineering would enable new communities of users to explore biology as a medium for creative expression. Utilizing commodity 3D printing, we fabricated multiple functional fluidic devices for miniaturizing DNA assembly, the foundational technology for synthetic biology, along with fluidic valves, the essential component for creating programmable devices. 3D print your own DNA assembler today!
miRNA-based Genetic Circuits for Cancer Detection
synbio | High-throughput library generation
miRNAs are a recently discovered class of nucleic acids involved in almost every aspect of human development. By querying only a small number of miRNAs in a cell, cell-type, and even disease state, can be discerned. In collaboration with MIT Professor Ron Weiss, we are developing tools for the high-throughput generation of libraries of genetic circuits to identify and destroy cancer cells via miRNA signatures. Additionally, with these circuit libraries we are cataloging expression profiles for every known human miRNA.
Rapid Protein Engineering for Bioremediation and Personalized Therapeutics
synbio | designer enzymes
A major bottleneck in protein engineering is the rate at which protein designs can be tested. We are developing fluidic platforms for the high-throughput synthesis and assay of hundreds of enzyme variants, in parallel, enabling protein engineers to rapidly test their designs to find the best candidates. We have utilized these platforms to prototype enzymes for the bioremediation of toxic chemicals and anti-viral therapeutics for personalized medicine.
Artificial Nose
synthetic organs
In my post-doctoral work, I served as a team leader for the $10 million DARPA-funded “RealNose” Project. In collaboration with Principal Research Scientist Shuguang Zhang, I demonstrated the first synthesis
of olfactory receptors in a microfluidic device for use with an odorant-detecting biosensor.
Microfluidic Gene Synthesis
synbio |automated DNA assembly
DNA synthesis is the foundational technology of synthetic biology. In my doctoral work, I demonstrated the first assembly of genes in a microfluidic, "lab-on-a-chip" system. I also developed platforms integrating gene synthesis with protein synthesis. Such miniaturized fluidic platforms are promising tools for the high-throughput automation of not only DNA assembly, but also the integration of protein and cellular engineering.
Ion- and Electron-beam Mediated Nanostructure Fabrication
nanotechnology
In my masters work I developed technologies for creating nanostructures utilizing energetic beams. I used a focused ion beam (FIB) to directly pattern nanoparticles composed of a variety of materials, and a technique for electrostatically attracting ionized inorganic nanoparticles to surfaces, termed nanoxerography.